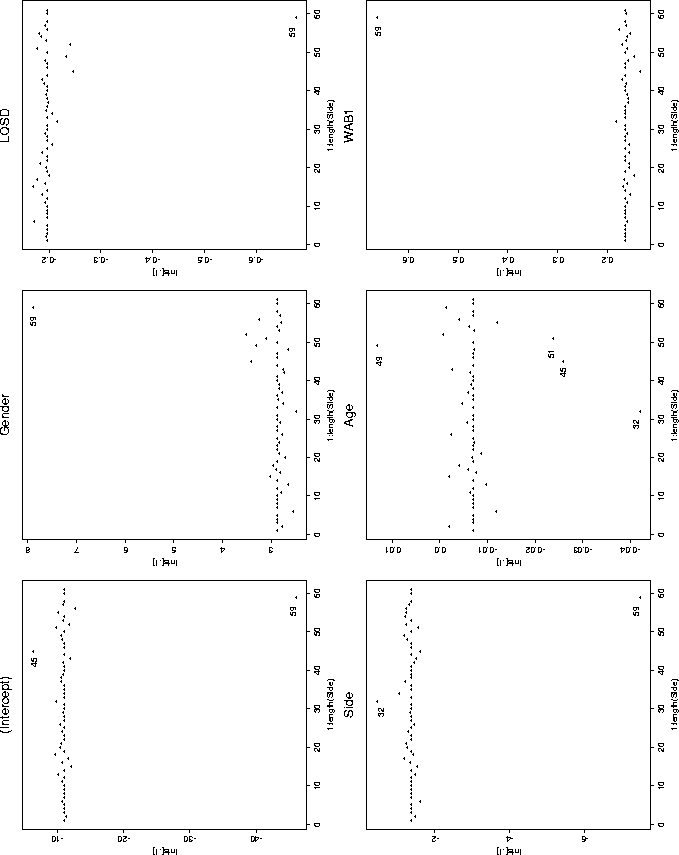
There are good measures of influence for glm models, but they don't seem to be implemented (i.e., I could not find them) in the standard packages. Rather than just give up, I quickly hacked together a ``Poor Man's'' influence function. The basic idea of the following code is to fit the glm model with each data point in turn removed. The summary is then a plot of each of the coefficients, with as many points on the plot as there are data points. This plot is surprisingly effective and quickly shows us where the pathology in the data lies.
# Heavily annotated, for class
# Generate a vector of all ones, of length the number of observations.
W <- rep(1,length(Side))
# Generate current weights.
WW <- W
# Fit the base model
mymod <- glm(Status ~ Gender + LOSD + Side + Age + WAB1, weight=WW,
family=binomial)
# Save the coefficients, just in case
base.coef <- coefficients(mymod)
# Set up a matrix to hold the coefficients for each model with one
# point deleted.
infs <- matrix(0, nrow=length(Side), ncol=length(coefficients(mymod)))
# Loop through the data, setting one weight equal to 0.
# Save the coefficients in the infs matrix
for (i in 1:length(Side)){
WW<-W; WW[i] <- 0;
infs[i,] <- coefficients(update(mymod, . ~ .))
}
## Can't think of an easy, smart summary, so just plot the
## coefficients.
#Set up a 2 x 3 plotting frame
par(mfrow=c(2,3))
#Do the 6 plots. After each plot, allow the user to
# identify the points which look interesting.
for(i in 1:6){
plot(1:length(Side), infs[,i])
title(names(coefficients(mymod))[i])
identify(1:length(Side), infs[,i])
}
After running that code, and interacting with the various plots, you end up with the following plot.

There are other things we could have plotted (like following the deviance as the models changed), but this plot alone tells us much of the story.