
Lets work on that, in that order (which is really back-to-front, but useful for teaching purposes).
The fitted values (on the probability scale) are obtained via,
402 > fitted(mymod)
1 2 3 4 5 6 7
0.2607849 0.1817866 0.1652202 0.09994062 0.07278457 0.06535816 0.06535816
8 9 10 11 12 13 14
0.06535816 0.05864156 0.05257638 0.04710711 0.04710711 0.04710711 0.04710711
15 16 17 18 19 20 21
0.03775043 0.03376847 0.02698616 0.02698616 0.02411117 0.02411117 0.0192299
22 23
0.01716665 0.01367159
The predicted values (on the logit scale) are,
402 > predict(mymod)
1 2 3 4 5 6 7
-1.041893 -1.50429 -1.619889 -2.197885 -2.544682 -2.660281 -2.660281
8 9 10 11 12 13 14
-2.660281 -2.77588 -2.891479 -3.007079 -3.007079 -3.007079 -3.007079
15 16 17 18 19 20 21
-3.238277 -3.353876 -3.585074 -3.585074 -3.700673 -3.700673 -3.931872
22 23
-4.047471 -4.278669
402 > fitted(mymod) - unlogit(predict(mymod))
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
(unlogit is the following simple function),
402 > unlogit <- function(x) { exp(x)/(1 + exp(x)) }
For communication purposes, it makes sense to work on the probability scale (and hence use fitted), but for all other purposes the predicted values are what we want. In particular, the predict function has an option that will give the Standard Errors of the predicted values (on the logit scale), viz.,
402 > predict(mymod,se=T)
$fit:
1 2 3 4 5 6 7
-1.041893 -1.50429 -1.619889 -2.197885 -2.544682 -2.660281 -2.660281
8 9 10 11 12 13 14
-2.660281 -2.77588 -2.891479 -3.007079 -3.007079 -3.007079 -3.007079
15 16 17 18 19 20 21
-3.238277 -3.353876 -3.585074 -3.585074 -3.700673 -3.700673 -3.931872
22 23
-4.047471 -4.278669
$se.fit:
1 2 3 4 5 6 7
0.7682786 0.5943294 0.556385 0.4298909 0.4298809 0.4441498 0.4441498
8 9 10 11 12 13 14
0.4441498 0.4647341 0.4908397 0.5216385 0.5216385 0.5216385 0.5216385
15 16 17 18 19 20 21
0.5942932 0.6348851 0.7222049 0.7222049 0.7682357 0.7682357 0.8637766
22 23
0.9129226 1.013299
$residual.scale:
[1] 1.189907
$df:
[1] 21
So, a 95% confidence interval for the first predicted value on the logit scale would be

Transferred to the probability scale, this is an interval
 , with a center at
, with a center at  .
.
But what about predictions nearer the take-off temperature?
predict(mymod, data.frame(Temp=c(30,31,32,33)), se=T)
$fit:
1 2 3 4
1.616887 1.501288 1.385689 1.27009
$se.fit:
1 2 3 4
1.972255 1.917739 1.863302 1.80895
(That was pretty sneaky, but the predict function requires a data frame predictor values).
The bottom line is that at  F our model extrapolates to
F our model extrapolates to
 , with a 95% confidence interval of
, with a 95% confidence interval of
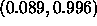 , which is VERY wide. Even with this wide interval,
the point estimate is something to worry about.
, which is VERY wide. Even with this wide interval,
the point estimate is something to worry about.